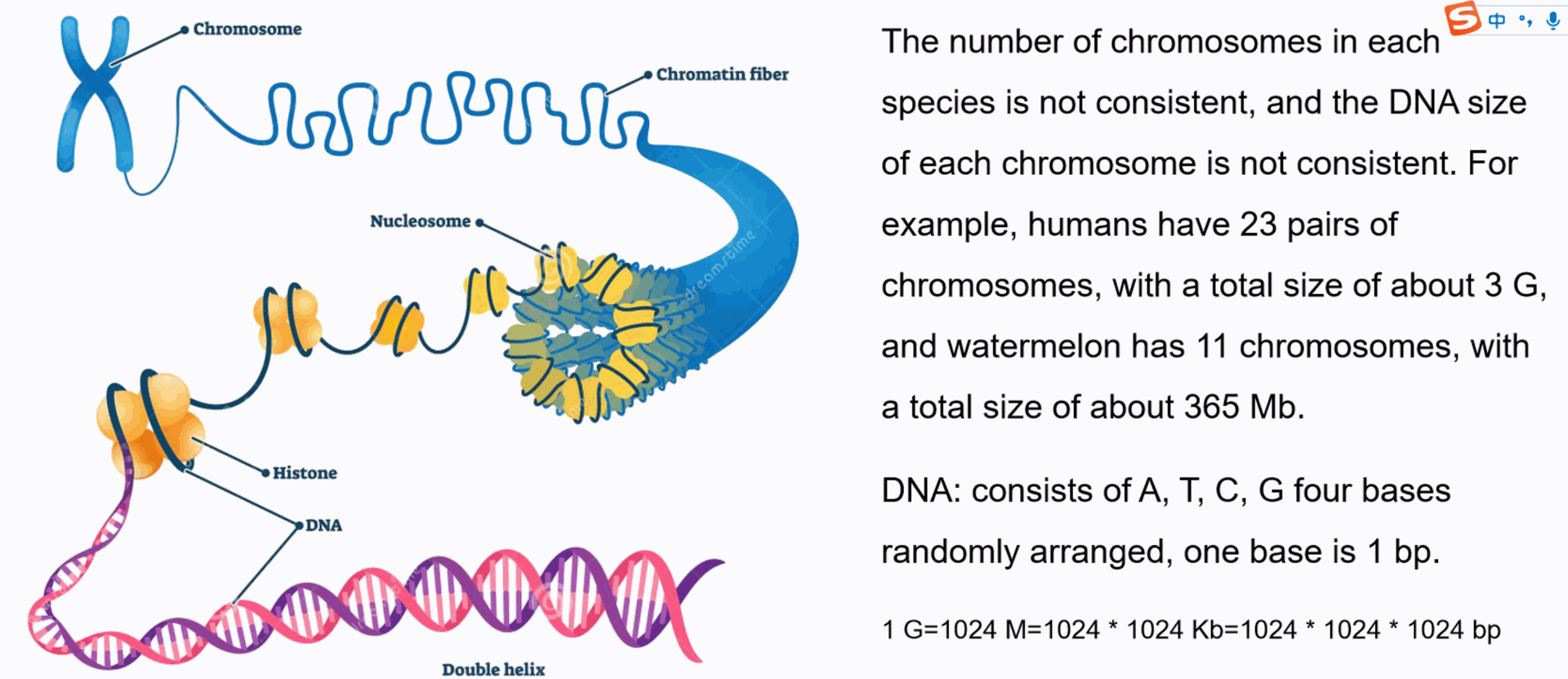
Various types of Sequencing
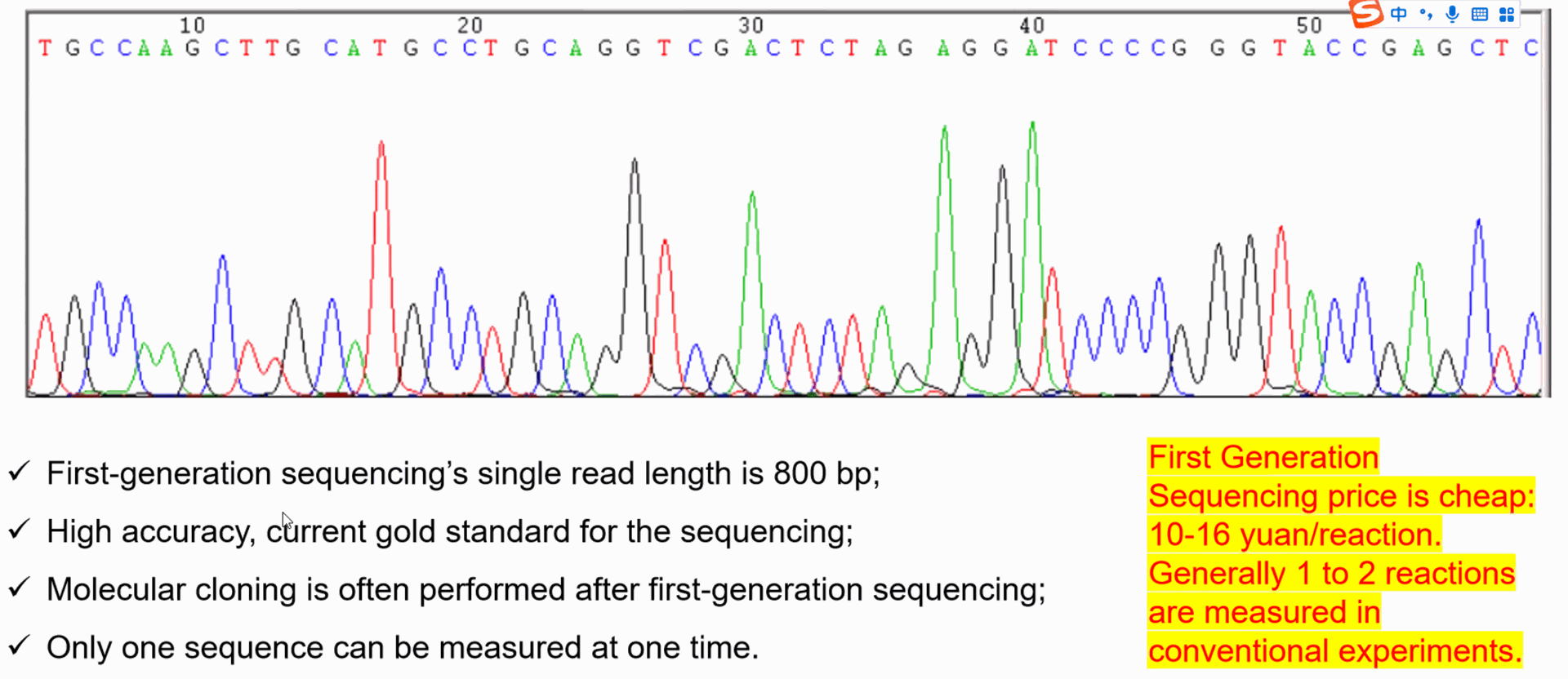
What is Sanger Sequencing?
Sanger Sequencing
- **Method**: Sanger sequencing, also known as chain termination or dideoxy sequencing, was the first widely used method for sequencing DNA. It involves copying DNA fragments and incorporating chain-terminating nucleotides that stop the DNA synthesis at specific points. These terminated fragments are then separated by size, and the sequence is determined by reading the order of the nucleotides.
- **Read Length**: Sanger sequencing produces relatively long reads (up to 800-1000 base pairs).
- **Accuracy**: It is highly accurate, with an error rate of around 0.001%.
- **Throughput**: Low throughput, making it more suitable for sequencing small regions of DNA, like individual genes.
- **Applications**: Used for small-scale projects, such as sequencing single genes, validating next-generation sequencing results, or detecting mutations.
What is NGS from Illumina?
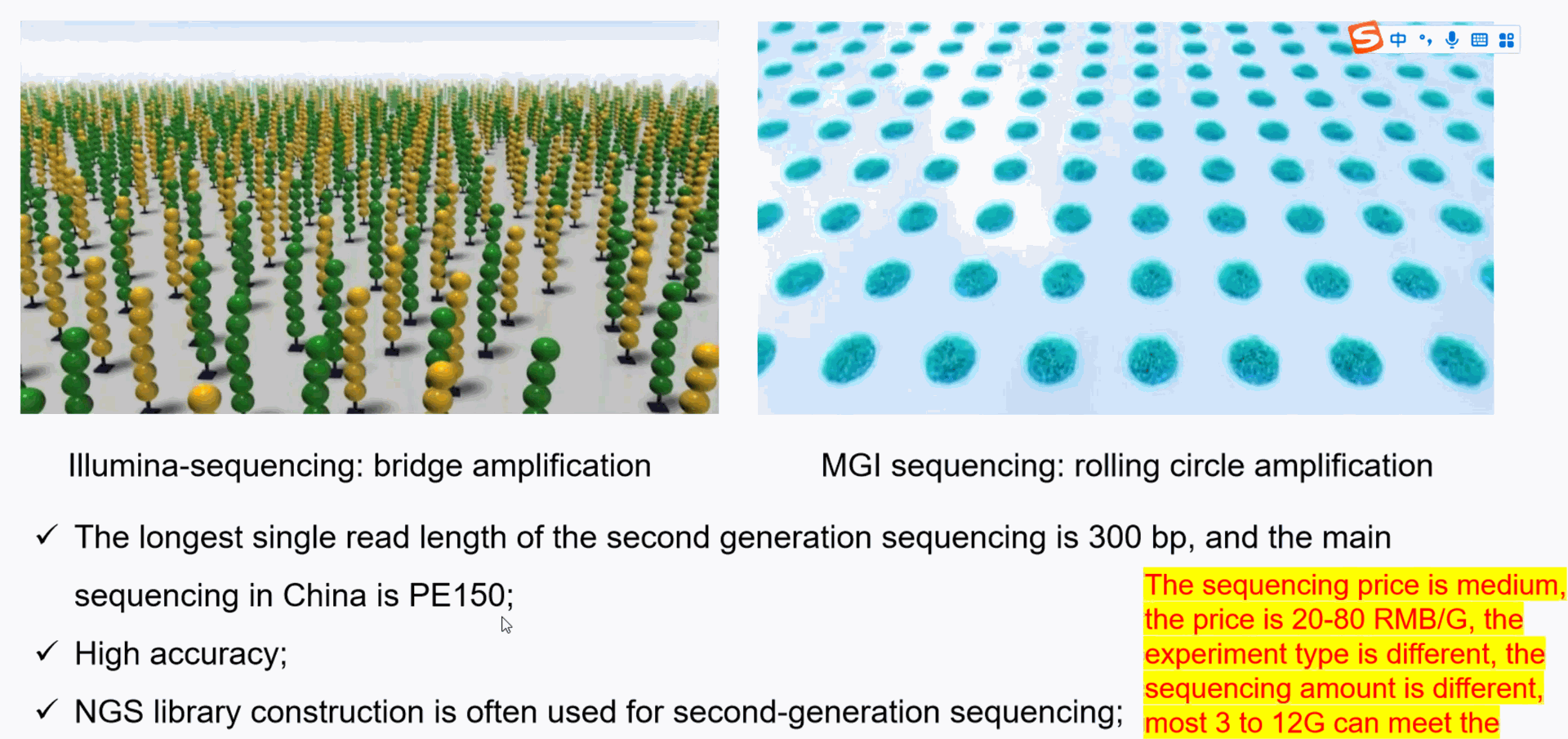
Illumina Sequencing
- **Method**: Illumina sequencing, also known as sequencing by synthesis (SBS), involves fragmenting DNA, attaching adapters, and binding the fragments to a flow cell. The fragments are then amplified to form clusters, and fluorescently labeled nucleotides are incorporated one by one during DNA synthesis. A camera captures the fluorescent signals to determine the sequence.
- **Read Length**: Short reads, typically 50-300 base pairs.
- **Accuracy**: High accuracy with an error rate of around 0.1% or lower.
- **Throughput**: Very high throughput, capable of sequencing billions of fragments simultaneously, making it suitable for large-scale projects like whole-genome sequencing.
- **Applications**: Used in genomics, transcriptomics, epigenomics, and large-scale projects requiring massive amounts of data, such as population studies and cancer research.
NGS explained from Oxford Nanopore and Pacbio
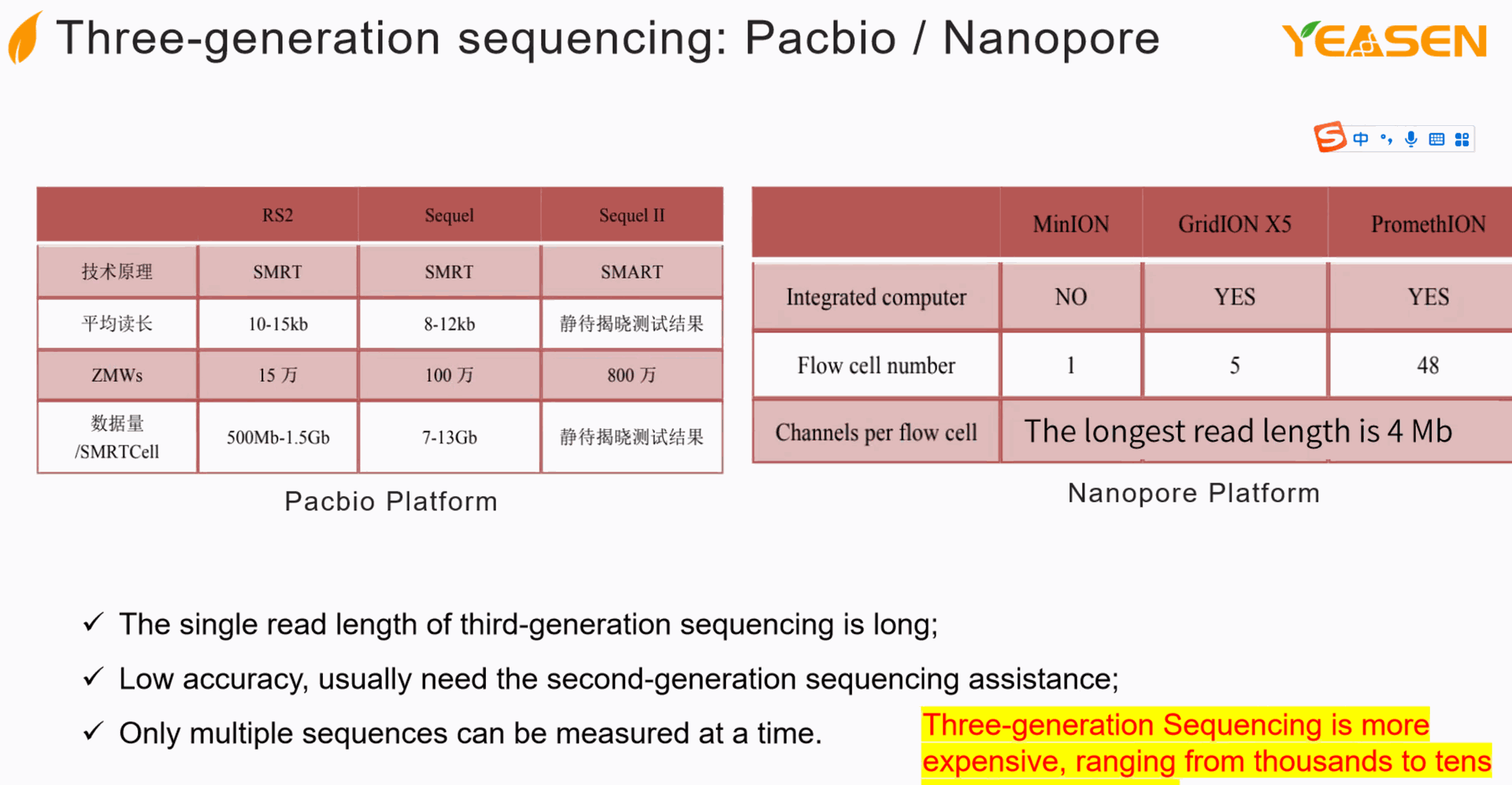
PacBio Sequencing (Pacific Biosciences)
- **Method**: PacBio sequencing uses Single Molecule Real-Time (SMRT) technology. DNA molecules are immobilized in tiny wells, where DNA polymerase synthesizes new DNA strands. Fluorescently labeled nucleotides are added one by one, and the system captures the light emitted when a nucleotide is incorporated.
- **Read Length**: Long reads, often exceeding 10,000 base pairs, with some reads reaching over 100,000 base pairs.
- **Accuracy**: The initial error rate is higher than Illumina (around 10-15%), but the use of multiple passes (circular consensus sequencing) can reduce this to around 0.1%.
- **Throughput**: Lower throughput than Illumina but sufficient for projects requiring long reads.
- **Applications**: Ideal for sequencing regions with complex structures, such as repetitive sequences, structural variants, and whole genomes of organisms with large or complex genomes.
### 4. **Oxford Nanopore Sequencing**
- **Method**: Oxford Nanopore sequencing involves passing a single strand of DNA through a nanopore (a tiny hole) embedded in a membrane. As the DNA passes through, it disrupts the ionic current, and these changes are used to determine the sequence in real-time.
- **Read Length**: Ultra-long reads, often exceeding 100,000 base pairs, with some reads surpassing 1 million base pairs.
- **Accuracy**: Lower than other methods, with a typical error rate around 5-15%, though improvements in algorithms have steadily increased accuracy.
- **Throughput**: Highly variable depending on the platform and flow cell used, ranging from moderate to high throughput.
- **Applications**: Particularly useful for sequencing long regions, identifying structural variants, assembling genomes, and real-time applications like pathogen detection and portable field sequencing.
What are the NGS applications in 2025?
Sanger sequencing, PacBio sequencing, Oxford Nanopore sequencing, and Illumina sequencing are all different methods of DNA sequencing, each with its own unique approach, advantages, and limitations.
Sequencing Key Differences
- - **Read Length**: Sanger and Illumina provide short to medium reads; PacBio and Oxford Nanopore offer long to ultra-long reads.
- - **Accuracy**: Sanger and Illumina are highly accurate, PacBio is accurate with post-processing, and Oxford Nanopore has lower initial accuracy but is improving.
- - **Throughput**: Illumina excels in high throughput, making it suitable for large-scale projects, while PacBio and Oxford Nanopore are more specialized for long-read sequencing needs.
- - **Applications**: Illumina dominates in large-scale genomics, PacBio and Oxford Nanopore are favored for complex genome assemblies and long-read applications, and Sanger is used for smaller-scale, high-accuracy sequencing tasks.
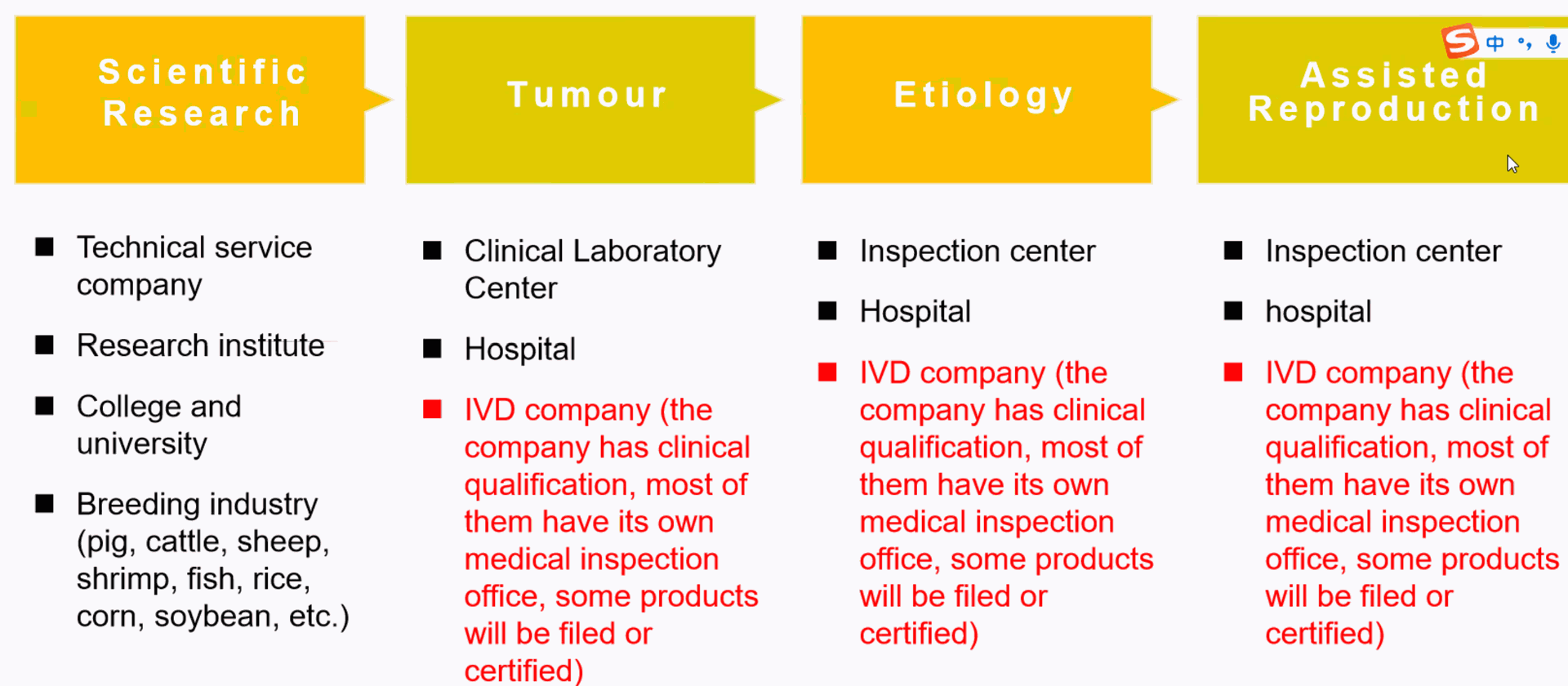
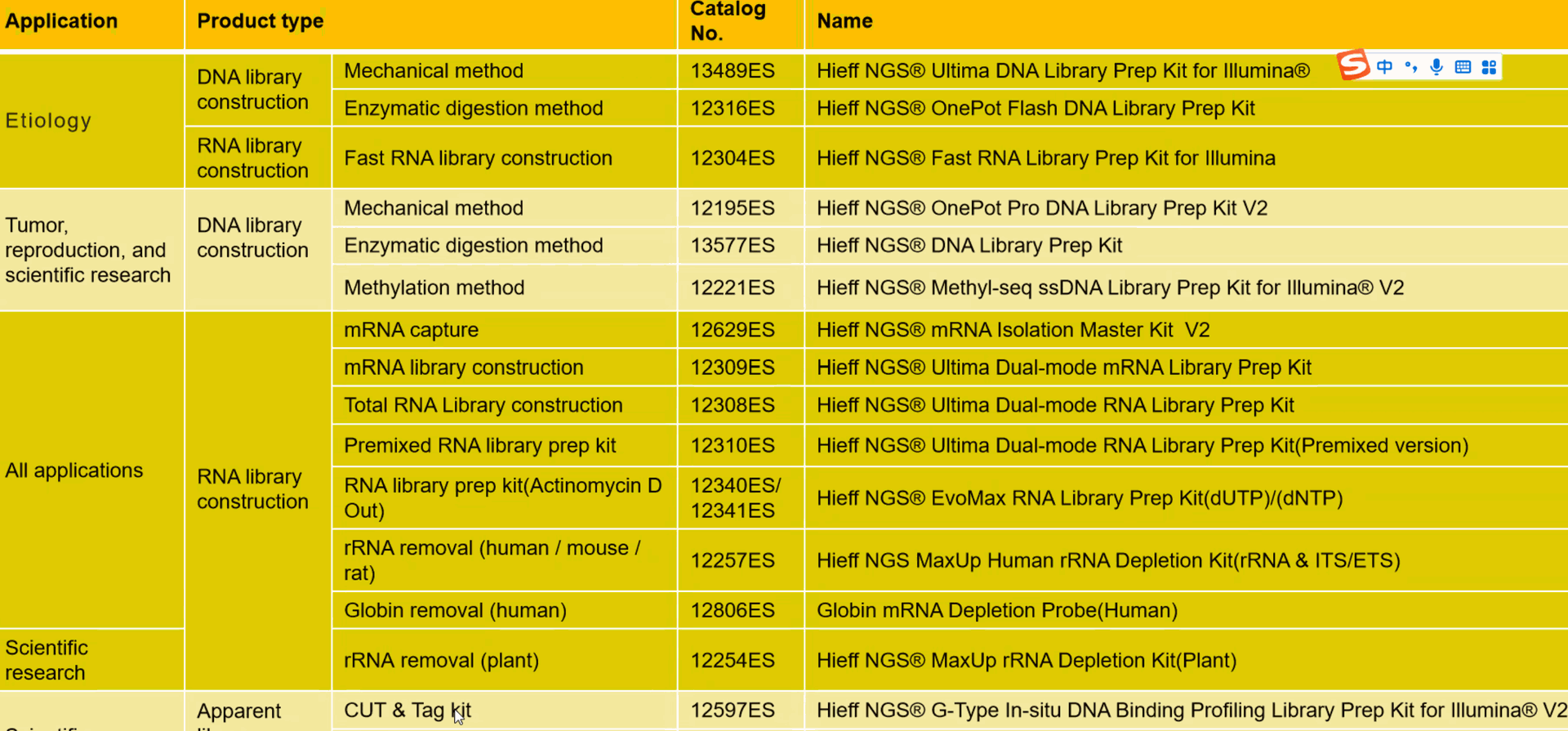
NGS what does it need?

Available kits for NGS
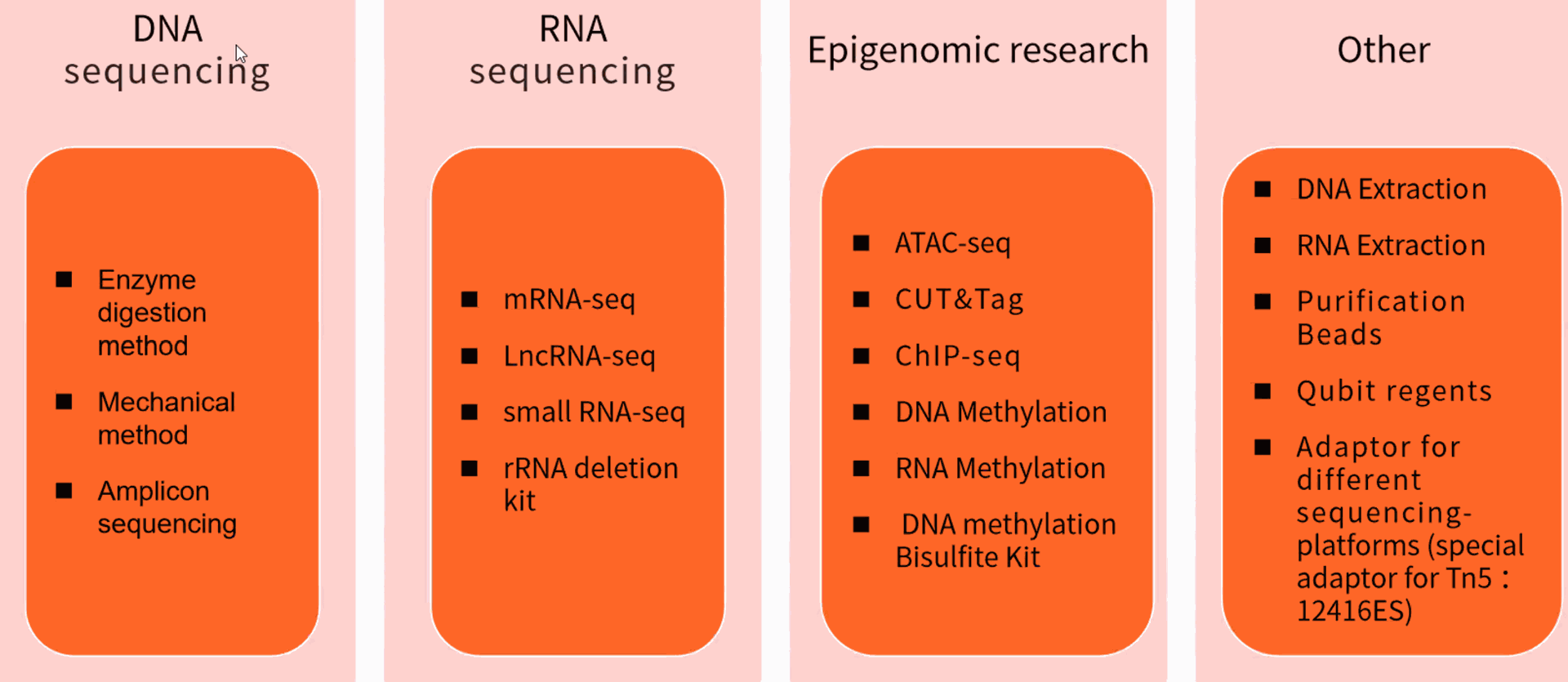
Different NGS sequencing fields of research
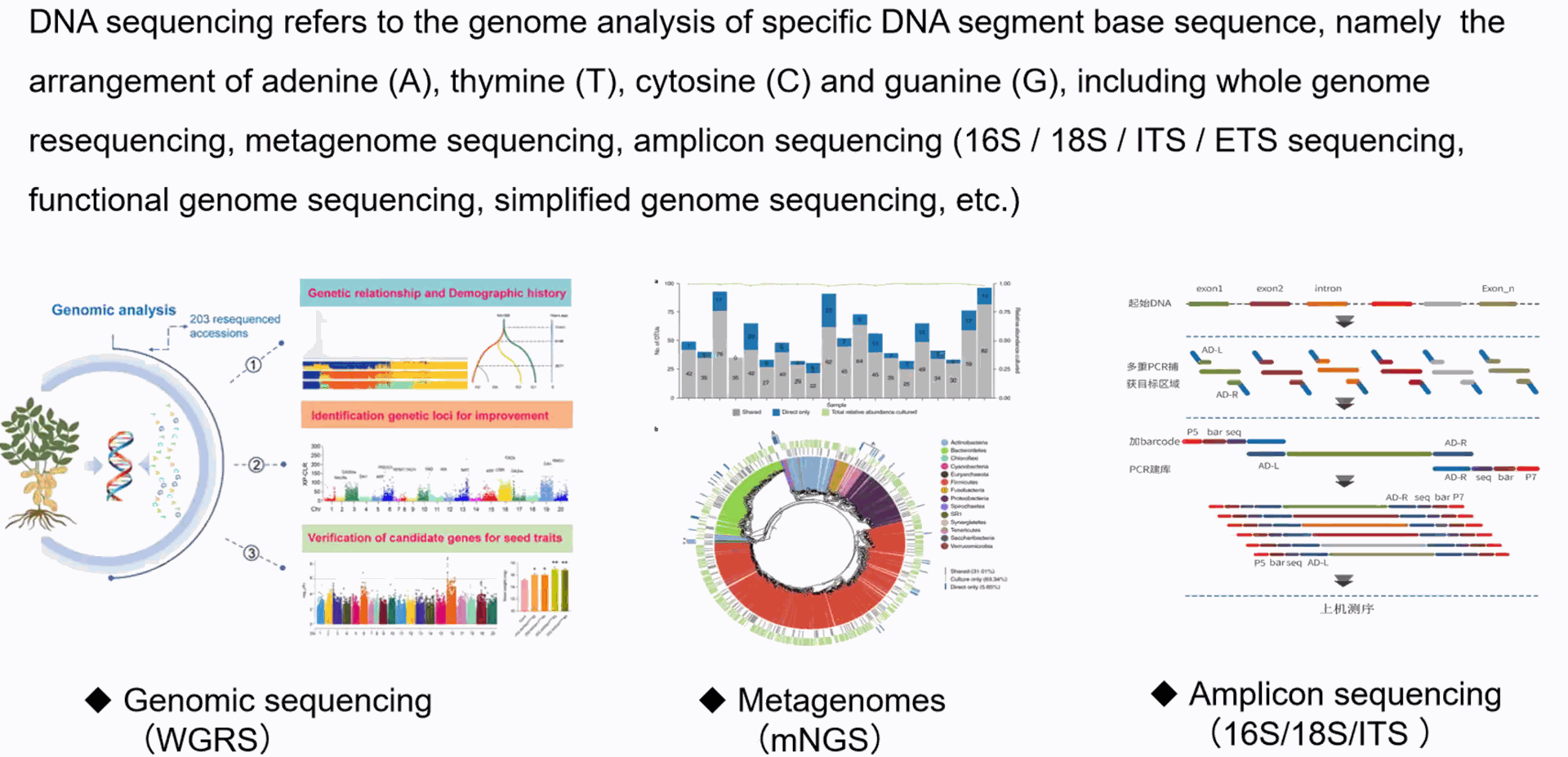
Metagenomes Amplicon sequencing
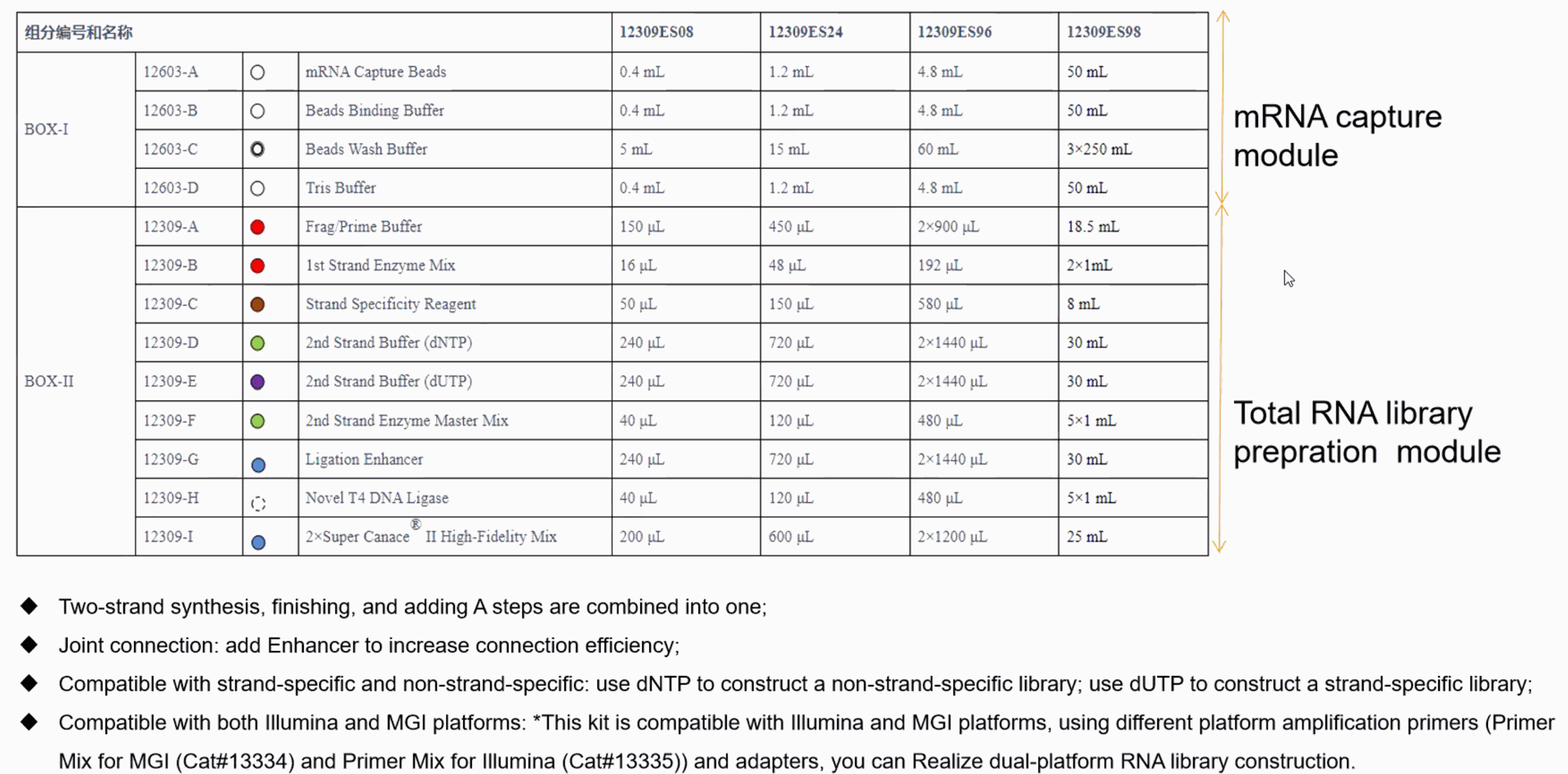
NGS Library construction kits
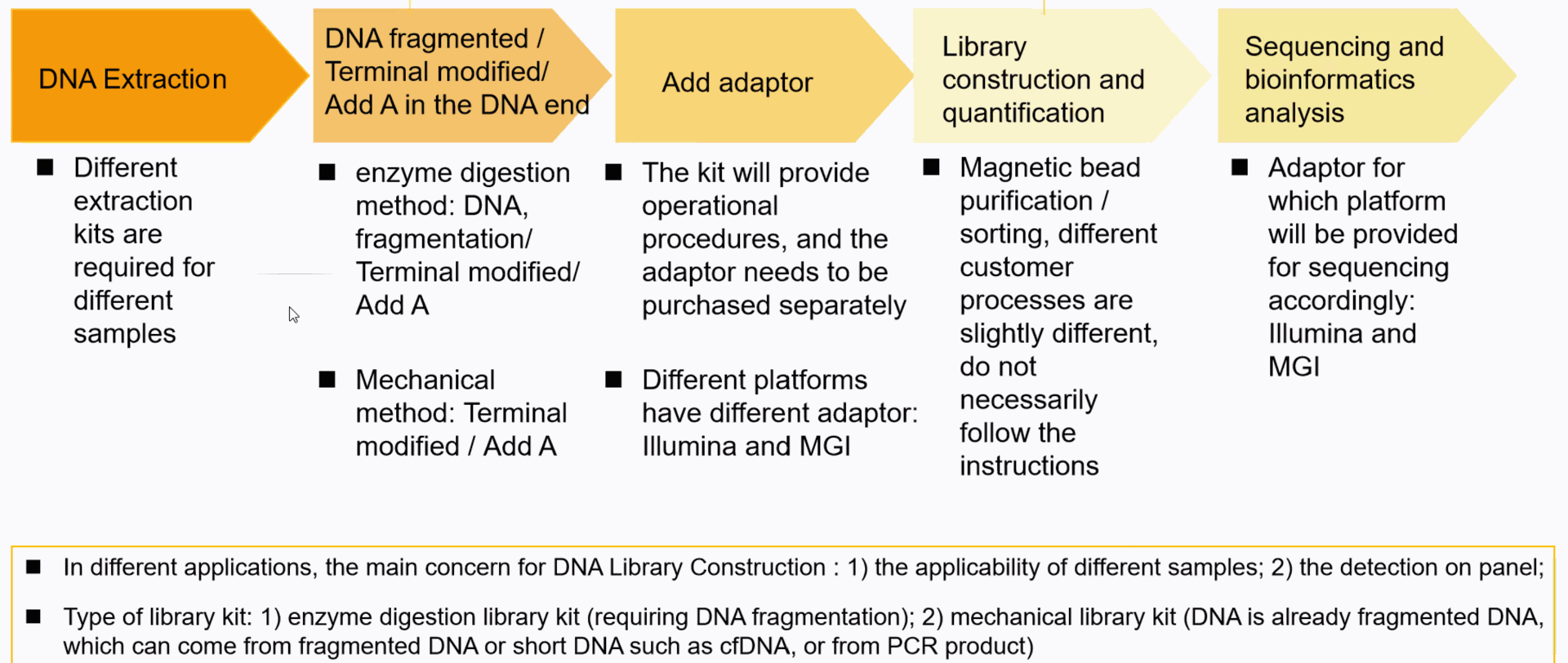
- magnetic beads in NGS: DNA\RNA\mRNA magnetic beads
- Key enzymes involved in NGS library construction
- Overview of T4 DNA Ligase
- DNase I and Their Applications in Biomedicine
- Qubit, what a powerful tool for NGS library quantification!
- How to Choose your NGS Adapters?
- About NGS-related technology, how much do you know?
- Have you learned how to build a PCR-Free library?
- DNase I hypersensitive sites-Important applications of DNase I
Start writing here...